Genes: New High Fashion on Campus?
The year 2001 marks the beginning of a new
era in which we know the entire genome sequence for several organisms,
including our own species, Homo sapiens. We are now
faced with making sense and use out of the vast assemblage of
information about our genome. Molecular biology
once pursued the understanding of gene structure and expression.
Now, the field of bioinformatics has the task of
organizing and understanding the design and evolution of the large databases we
know as genomes. The sheer vastness of the data will require new
approaches to handling databases and accessing the data therein. This
opportunity to develop new technology and methodology might be likened to the
challenge of landing a human on the moon.
One approach will be to
compare our genome architecture to that of closely and distantly related
 species - not comparing gene structure but rather the
clusters and orderings of genes on the chromosomes on which they lie.
The evolution of gene patterns may teach us about the function of the
large amount of "junk" DNA that lies between the transcribed DNA that codes for
proteins and structural RNAs. The Biology Department has joined the
cadres of scientists who will be exploring genome organization in a variety of
species.
species - not comparing gene structure but rather the
clusters and orderings of genes on the chromosomes on which they lie.
The evolution of gene patterns may teach us about the function of the
large amount of "junk" DNA that lies between the transcribed DNA that codes for
proteins and structural RNAs. The Biology Department has joined the
cadres of scientists who will be exploring genome organization in a variety of
species.
In 1998 The University of Massachusetts Automated DNA Sequencing
Facility was established with Biology faculty member Ron Adkins as
Director.
Since its opening, the facility has applied its machinery to the
sequencing of DNA of over 200 different species. The facility services
the sequencing needs of 60 laboratories in the 5-College community.
Each year sees its usage increasing with a 6,400 sequence throughput in the
past year, representing 3.2 million nucleotide pairs identified.
Facility director Ron Adkins is interested in the deep structure of
mammalian phylogenetic divergence as exemplified by this tree of mammalian
evolutionary branching, excerpted from one of his earlier publications, which
includes primates, tree shrews (Tupaia), flying lemurs
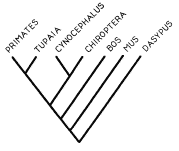 (Cynocephalus), bats (Chiroptera), mice (Mus), cows (Bos) and armadillos (Dasypus). Ron's current research is exploring the evolution
of the deep structure of the speciose group of mammals which includes
mice and rats. Another Biology faculty member
Yin-Long Qiu
uses the facility to study plant genomics.
Microbiology Professor Derek Lovley used the facility extensively to
contribute to the Geobacter genome project. Biology courses use
the facility to provide training in the application of the tools of molecular
biology.
(Cynocephalus), bats (Chiroptera), mice (Mus), cows (Bos) and armadillos (Dasypus). Ron's current research is exploring the evolution
of the deep structure of the speciose group of mammals which includes
mice and rats. Another Biology faculty member
Yin-Long Qiu
uses the facility to study plant genomics.
Microbiology Professor Derek Lovley used the facility extensively to
contribute to the Geobacter genome project. Biology courses use
the facility to provide training in the application of the tools of molecular
biology.
In addition to genomics research per se, the facility is being
used by university geneticists in a variety of ways. Geneticist
Randy Phillis explains typical scenarios: "Once
we have induced a mutation in our organism of choice, we can identify the
locus of the mutation and sequence the gene. Then we can compare that
gene to the worldwide database of gene sequences and perhaps find homologous
genes whose properties have been identified previously. This gives us
clues to how our gene works. Second, we can engineer a mutation
in a known gene in order to explore the effects of such a mutation. The
sequencing facility can quickly allow us to check that our engineering
created the mutation we desired."
The Sequencing Facility is an indicator of the serious commitment the
University and its biology faculty have made to support modern genomics
research and teaching.