In our first year at UMass Amherst we have published independent projects with our collaborators confirming two of our most key basic findings. We have also published two reviews and one technology review. Finally we were able to acquire two major grants including NIH R00 and UM1 team grant through the 4D Nucleome consortium. Highlights from our 2020 publications below.
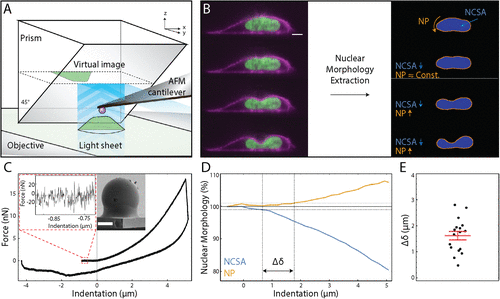
Major finding 1, Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus (initial publication Stephens et al., 2017 MBoC). Working with Physicist at the University of North Carolina Chapel hill we confirmed this novel finding by using an advanced atomic force microscope coupled with single plan illumination microscopy (AFM-SPIM). The ability to track extension/compression respectively with micromanipulation and AFM-SPIM provides the unique ability to separate the relative contributions of chromatin and lamin A to different regimes. Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics (Hobson et al., 2020 MBoC). TAKE HOME POINT: Chromatin dominates the initial short extension/compression while lamin A controls the high extension/compression regime via strain stiffening of the cell nucleus.
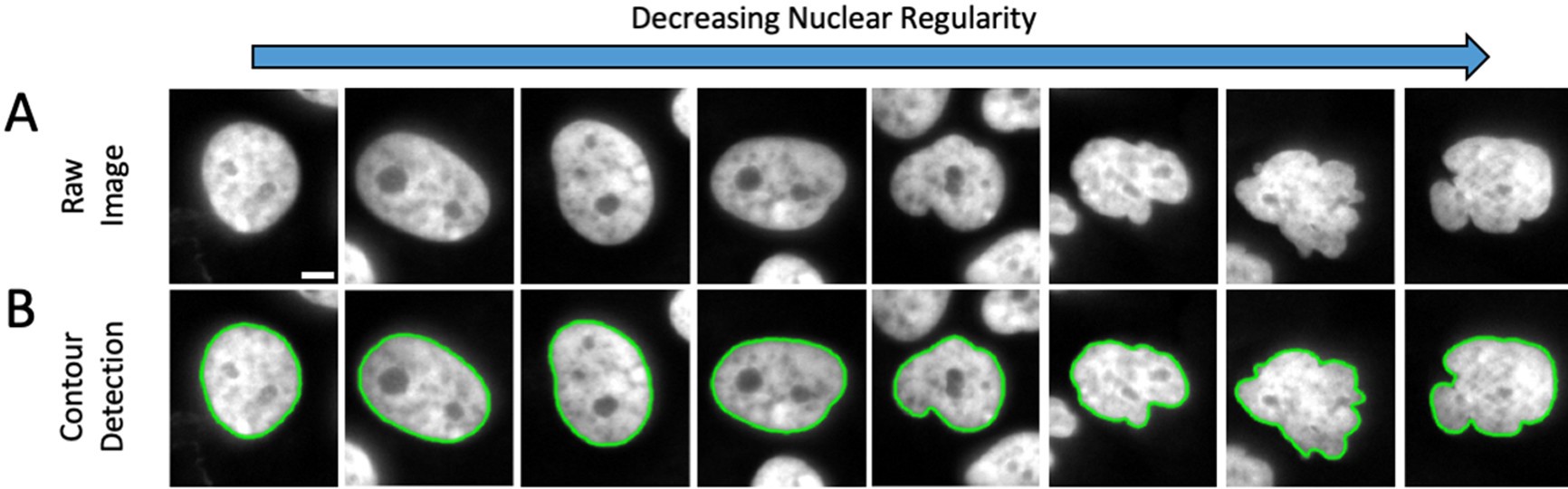
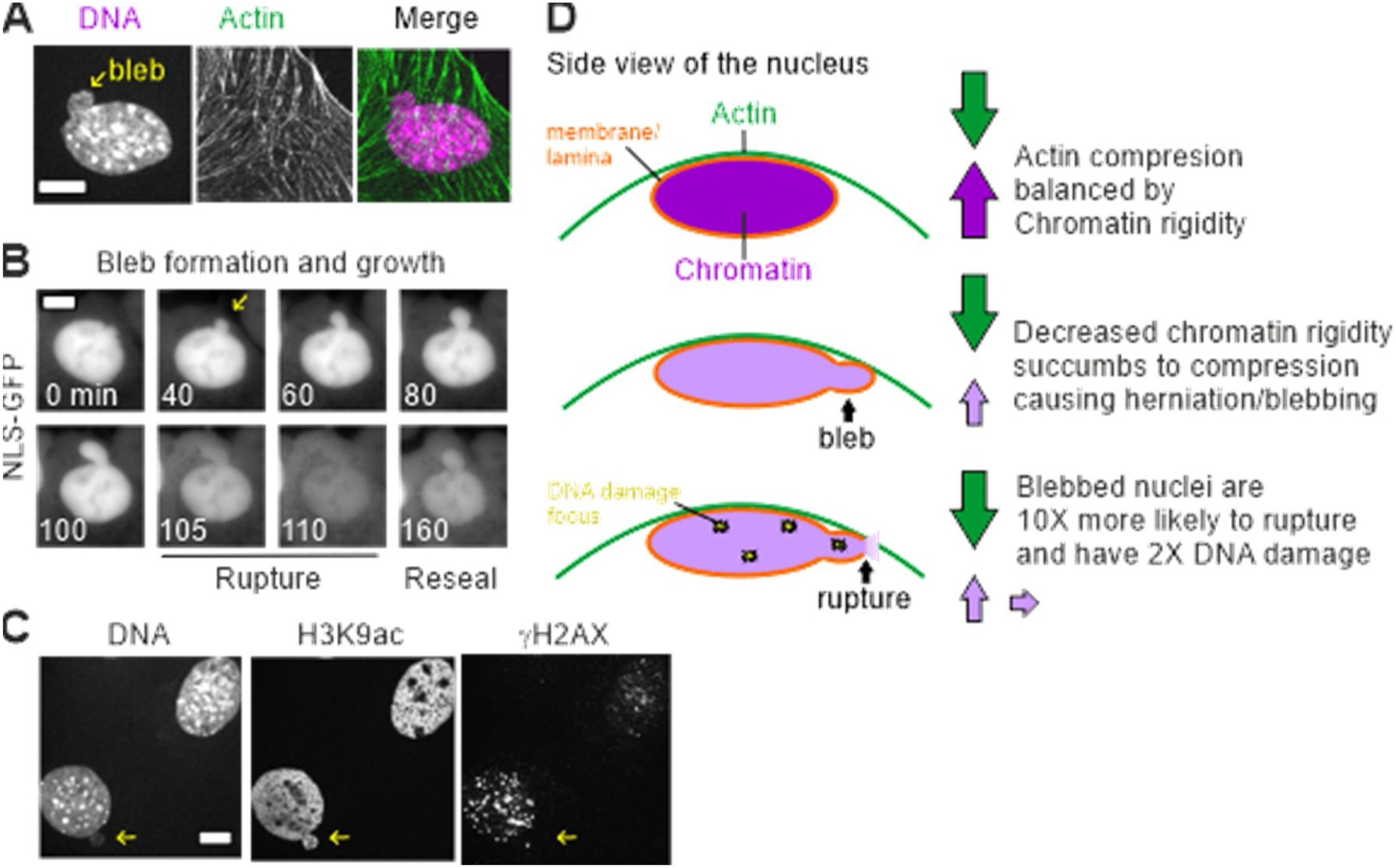
Major finding 2, chromatin, DNA and the proteins that organize and compact it, can control nuclear morphology/shape. Abnormal nuclear morphology is a hallmark of human diseases and recently has been tied to loss of nuclear function (see our review below). Our initial finding, Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins (initial publication Stephens et al., 2018 MBoC). In collaboration with the Lele lab (now at Texas A&M) a gene screen of chromatin proteins affect on nuclear shape revealed many are essential for nuclear shape maintenance, entitled High-throughput gene screen reveals modulators of nuclear shape (Tamashunas et al., 2020 MBoC). TAKE HOME POINT: Our work, in conjunction with others that have inspired us, continues to show that chromatin and its mechanical contribution control key aspects of nuclear shape maintenance essential for healthy cells.
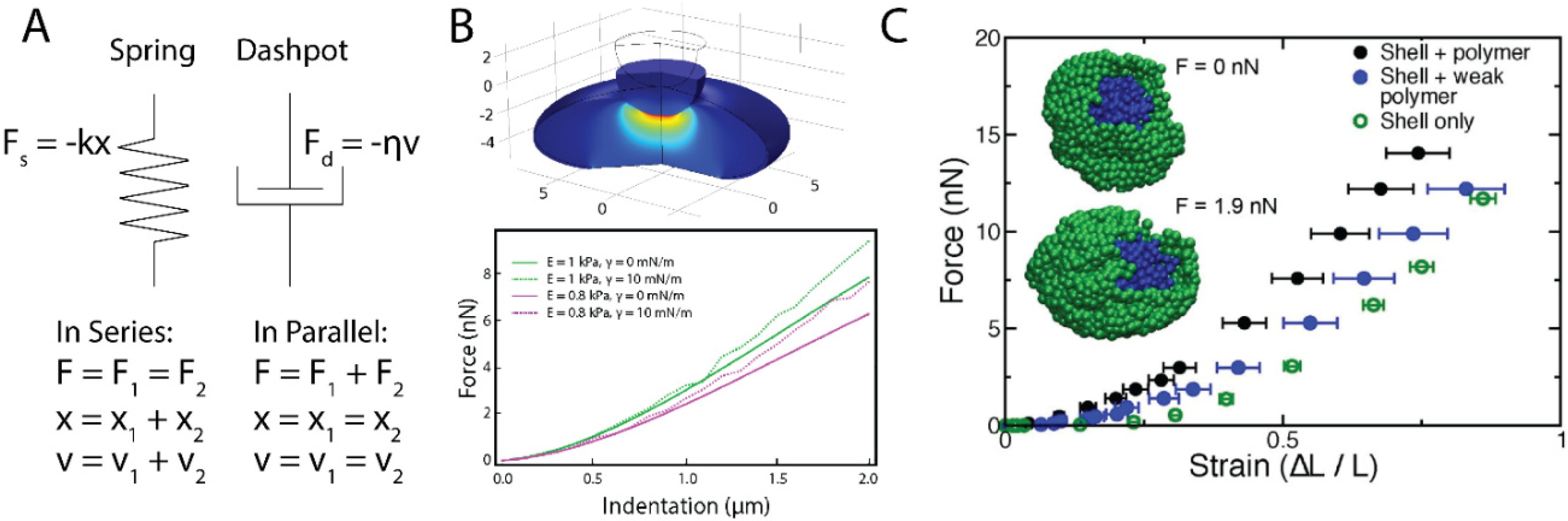
During our first year we also published two reviews meant to aid our young field as it grows. The first, Chromatin rigidity provides mechanical and genome protection (Stephens et al., 2020 Mut Res) details the above importance of chromatin's role in nuclear mechanics and morphology maintenance to the function of the cell nucleus. The second, Modeling of Cell Nuclear Mechanics: Classes, Components, and Applications (Hobson and Stephens et al., 2020 Cells) provides the first of its kind review of physical simulation modeling of the cell nucleus. We describe the three types of models along with their strengths and weakness, where we note that schematic models (spring and dashpot) are one dimensional, rely on idealized sweeping assumptions, and are in large part a restatement of the measured biophysical parameters providing no further insights. In both aspects of mechanical protection and modeling the field focus heavily on lamins but our research and many others detail the essential and, in cases, dominant role of chromatin as a mechanical protector of the nucleus and genome. TAKE HOME POINT: It is vital that we properly consider the contributions of both chromatin and lamins within functional experimental studies as well as in modeling the cell nucleus.
Finally, we were proud to be included on a technology review of new experimental chromatin measurements with many early investigators. Advances in Chromatin and Chromosome Research: Perspectives from Multiple Fields (Agbleke et al., 2020 Mol Cell). This team was lead by Andrew Seeber from Harvard and compiles a fantastic list of new methodology used to make advancements in chromatin biology. TAKE HOME POINT: Young scientists have generated many new technologies advancing the field of chromatin biology.
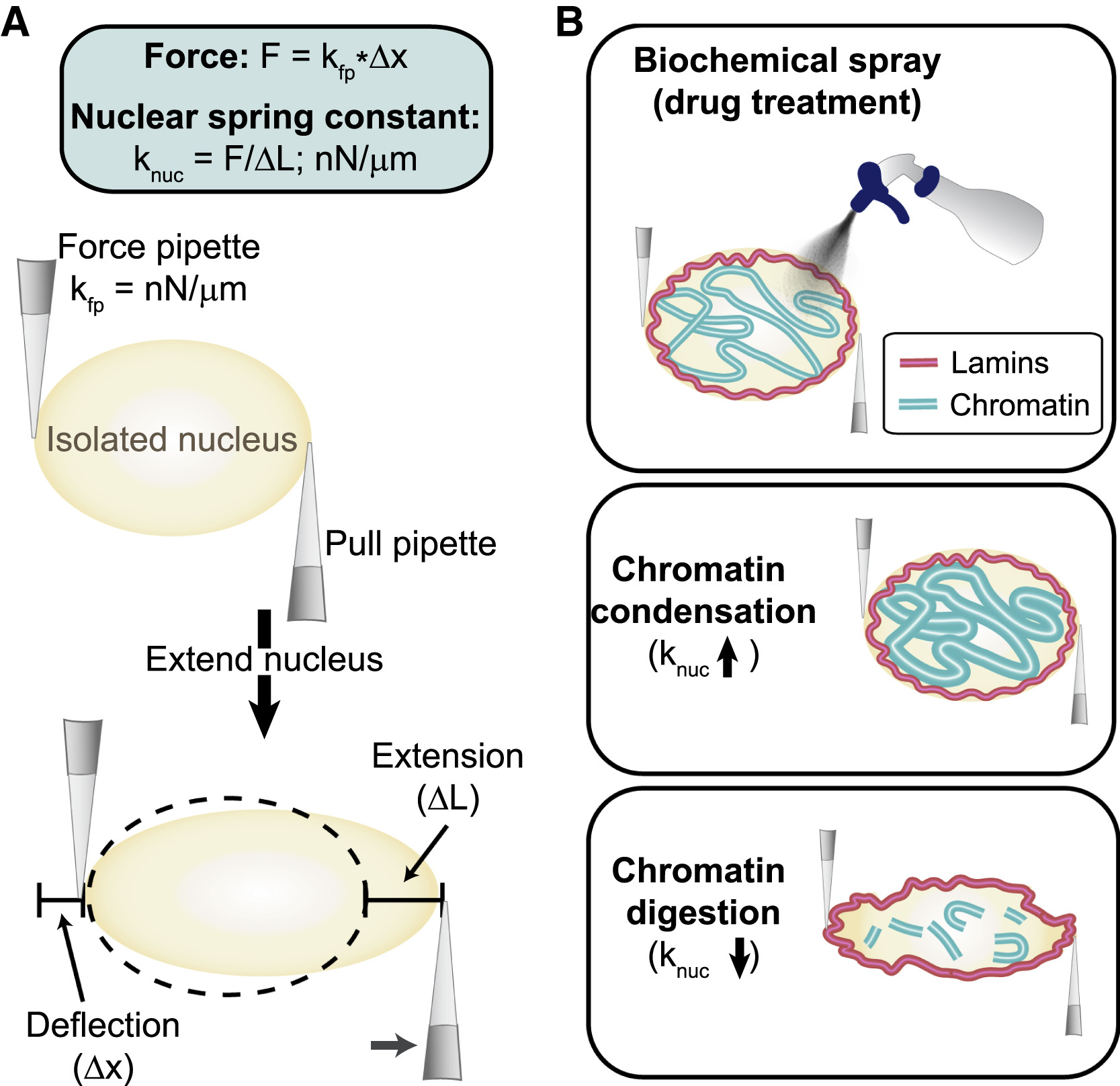
(A) Micromanipulation force measurement schematic of how the technique works. A force pipette (fp) and a pull pipette (pp) are used to extend the nucleus (nuc).
(B) A single isolated nucleus can be measured before and after biochemical treatment, providing fine-tuned control over nuclear chromatin mechanical and imaging measurements. The nuclear spring constant (knuc) increases during chromatin condensation and decreases after chromatin digestion.
