Research
Overview
The goal of the research in the Lopes lab is to understand the transcriptional regulation of phospholipid biosynthesis in yeast, and how phospholipid biosynthesis is coordinated with other biological processes. To address this goal, we are carrying on two inter-related projects. One project is designed to determine how phospholipid biosynthesis is coordinated with other biological processes via a set of transcription factors that belong to the basic helix-loop-helix (bHLH) family. Another project is focused on the regulation of the only essential phospholipid biosynthetic gene (PIS1) required for the synthesis of phosphatidylinositol (PI). This project is driven by the fact that virtually nothing is known about the transcriptional regulation of the PIS1 gene except that it is not coordinated with expression of the other phospholipid biosynthetic genes.
Combinatorial regulation of gene expression by basic helix-loop-helix proteins
Regulation of yeast phospholipid biosynthesis requires the Ino2p:Ino4p activators, which belong to the bHLH family of transcription factors. Initially, we were focused on the regulation of the INO2 and INO4 genes and found that INO2 expression is auto-regulated in a pattern that is identical to the regulation of the Ino2p target genes. More recently, we also carried out genetic analyses of these genes to define their roles in transcriptional regulation.
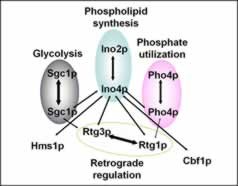
Fig. 1. Yeast bHLH protein interaction map.
Ovals denote previously known dimers and the processes they regulate. Arrows between ovals denote novel interactions. Broken arrow indicates a weak interaction.
Currently, we have turned our attention to how phospholipid biosynthesis is coordinated with other biological processes via the bHLH proteins. Yeast has nine bHLH proteins that regulate several different processes including: phosphate utilization, retrograde response (control of nuclear genes in response to mitochondrial damage), and glycolysis (Fig. 1). Since bHLH proteins are known to form multiple dimer combinations in other organisms, we reasoned that dimer partner selection could be a mechanism for coordination of the biological processes regulated by yeast bHLH proteins. Thus, we defined the yeast bHLH protein interaction map using the yeast two-hybrid system and co-purification of epitope-tagged recombinant proteins. As we predicted, this map identifies several novel bHLH dimer combinations (Fig. 1). In addition, since specific growth conditions affect expression of specific bHLH genes, then different growth conditions would be expected to change the bHLH protein interaction map. We are presently testing this prediction.
We are also determining the role of the yeast bHLH proteins in coordination of different biological processes using well-characterized candidate target genes for each biological process. To complement the candidate gene approach, microarray analysis is being used to define the target genes for each bHLH protein under conditions that affect the expression of the different bHLH encoding genes. Lastly, to define the mechanism(s) whereby bHLH dimers coordinate gene expression, we are determining the autoregulatory circuits that control expression of each bHLH-encoding gene.
Genomic analysis of PI synthesis in yeast
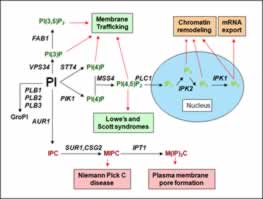
Recently, we have also focused our attention on the regulation of PIS1 expression The PIS1 gene is required for the synthesis of PI. PI and its metabolites (inositol polyphosphates, phosphoinositides, and sphingolipids) play important roles in a myriad of processes, including: chromatin remodeling, export of mRNA from the nucleus, vesicle trafficking, and signal transduction (Fig. 2). In spite of the importance of PI to the cell, relatively little is known about how its levels are regulated, particularly at the level of transcription of the PIS1 gene. Promoter deletion analysis reveals that the PIS1 promoter includes five regulatory elements: three upstream activation sequences (UAS) and two upstream repression sequences (URS). Our published studies reveal the carbon source regulates PIS1 expression through the URSGLY element and probably via a novel regulatory cascade. We have also determined that oxygen regulates PIS1 expression through the Rox1p repressor protein and the URSROX element. Screening a yeast gene deletion set, we identified 120 genes that affect expression of a PIS1 reporter. These genes suggest that processes such as peroxisomal function and DNA repair are coordinated with PI synthesis.
The knowledge that PI metabolites have roles that affect gene expression suggests that regulation of PI synthesis may affect several cellular processes. We are testing this directly by determining the phenotype of strains with altered PI synthesis and by using microarray hybridization to identify genes that are regulated in response to PI levels.