Publications
Nuclear blebs are associated with destabilized chromatin packing domains. Pujadas Liwag EM, Acosta N, Almassalha ML, Su Y, Kanemaki MT, Stephens AD, Backman V. bioRxiv 2024.03.28.587095
Nuclear shape is affected differentially by loss of lamin A, lamin C, or both lamin A and C. Pho, M; Berrada, Y; Gunda, A; Stephens, AD (2024). microPublication Biology. 10.17912/micropub.biology.001103.
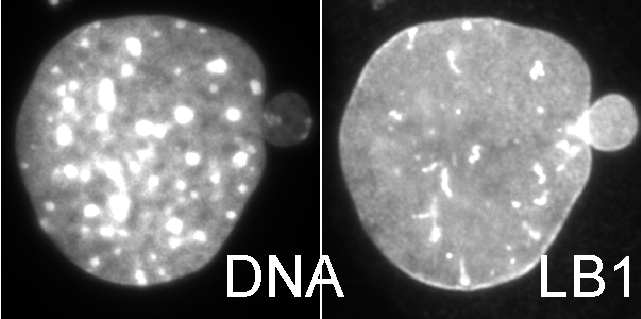 DNA density is a better indicator of a nuclear bleb than lamin B loss.Bunner S*, Prince K*, Srikrishna K*, Pujadas E*, Pellegrino P, Aiello J, McCarthy AA, Kuklinski A, Lawlor C, Jagtap S, Eweka I, Eastin E, Yas G, LaPointe N, Jackson O, Chen J, Schramm von Blucher I, Hardy J, Backman V, Janssen A, Packard M, Dorfman K, Almassalha L, Bahiru MS, Stephens AD. (2024) bioRxiv 2024.02.06.579152
DNA density is a better indicator of a nuclear bleb than lamin B loss.Bunner S*, Prince K*, Srikrishna K*, Pujadas E*, Pellegrino P, Aiello J, McCarthy AA, Kuklinski A, Lawlor C, Jagtap S, Eweka I, Eastin E, Yas G, LaPointe N, Jackson O, Chen J, Schramm von Blucher I, Hardy J, Backman V, Janssen A, Packard M, Dorfman K, Almassalha L, Bahiru MS, Stephens AD. (2024) bioRxiv 2024.02.06.579152
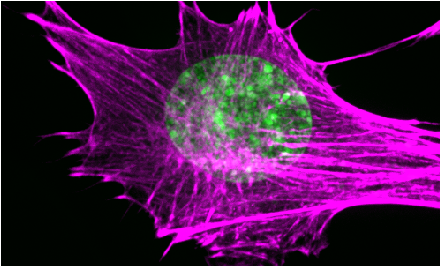 Actin contraction controls nuclear blebbing and rupture independent of actin confinement. Pho M, Berrada Y, Gunda A, Lavallee A, Chiu K, Padam A, Currey ML, Stephens AD. (2024) Mol Biol Cell. Feb 1;35(2):ar19. doi: 10.1091/mbc.E23-07-0292. PDF version here.
Actin contraction controls nuclear blebbing and rupture independent of actin confinement. Pho M, Berrada Y, Gunda A, Lavallee A, Chiu K, Padam A, Currey ML, Stephens AD. (2024) Mol Biol Cell. Feb 1;35(2):ar19. doi: 10.1091/mbc.E23-07-0292. PDF version here.
T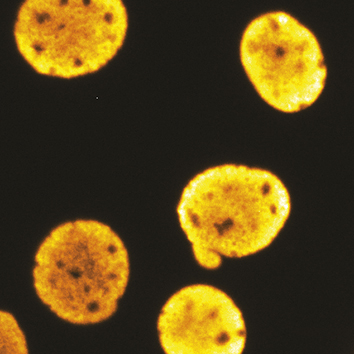 ranscription inhibition suppresses nuclear blebbing and rupture independent of nuclear rigidity.Berg IK*, Currey ML*, Gupta S, Berrada Y, Nguyen Viet B, Pho M, Patteson AE, Schwarz JM, Banigan EJ, Stephens AD. (2023) J Cell Sci 15;136(20):jcs261547. doi: 10.1242/jcs.261547. PDF version here.
ranscription inhibition suppresses nuclear blebbing and rupture independent of nuclear rigidity.Berg IK*, Currey ML*, Gupta S, Berrada Y, Nguyen Viet B, Pho M, Patteson AE, Schwarz JM, Banigan EJ, Stephens AD. (2023) J Cell Sci 15;136(20):jcs261547. doi: 10.1242/jcs.261547. PDF version here.
*** JCS Research Highlight Inhibiting transcription makes nuclear blebs fiction
C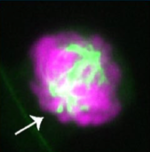 TCF is essential for proper mitotic spindle structure and anaphase segregation. Chiu K*, Berrada Y*, Eskndir N*, Song D, Fong C, Naughton S, Chen T, Moy S, Gyurmey S, James L, Ezeiruaku C, Capistran C, Lowey D, Diwanji V, Peterson S, Parakh H, Burgess A, Probert C, Zhu A, Anderson B, Levi N, Gerlitz G, Packard MC, Dorfman KA, Bahiru MS, Stephens AD. (2023) Chromosoma https://doi.org/10.1007/s00412-023-00810-w . PDF version here.
TCF is essential for proper mitotic spindle structure and anaphase segregation. Chiu K*, Berrada Y*, Eskndir N*, Song D, Fong C, Naughton S, Chen T, Moy S, Gyurmey S, James L, Ezeiruaku C, Capistran C, Lowey D, Diwanji V, Peterson S, Parakh H, Burgess A, Probert C, Zhu A, Anderson B, Levi N, Gerlitz G, Packard MC, Dorfman KA, Bahiru MS, Stephens AD. (2023) Chromosoma https://doi.org/10.1007/s00412-023-00810-w . PDF version here.
*** preLights written by Saanjbati Adhikari
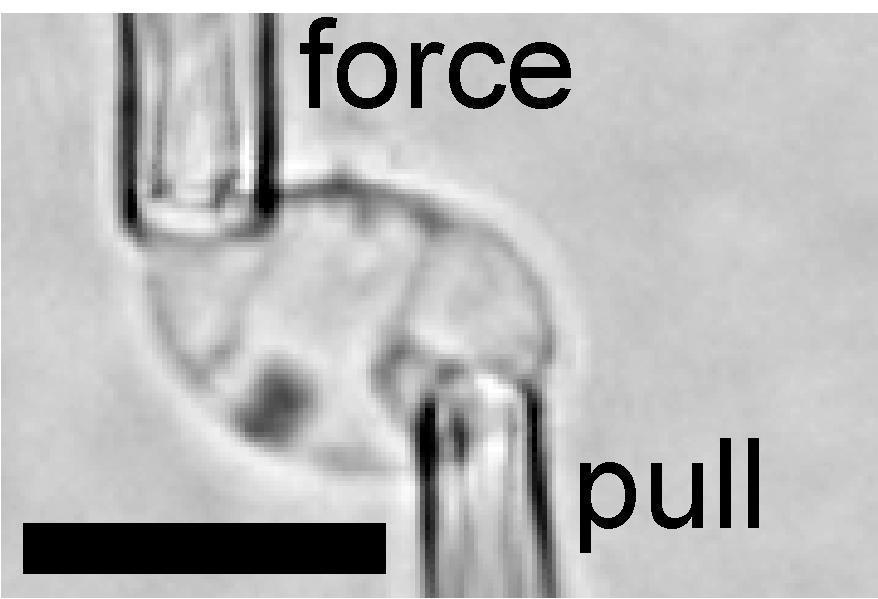 A versatile micromanipulation apparatus for biophysical assays of the cell nucleus. Currey ML, Kandula V, Biggs R, Marko JF, Stephens AD. (2022) Cell Mol Bioeng., 15, 303–312. PDF version here.
A versatile micromanipulation apparatus for biophysical assays of the cell nucleus. Currey ML, Kandula V, Biggs R, Marko JF, Stephens AD. (2022) Cell Mol Bioeng., 15, 303–312. PDF version here.
Mechanics and functional consequences of nuclear deformations. Kalukula Y, Stephens AD, Lammerding J, Gabriele S. (2022) Nat Rev Mol Cell Biol. 23, pages583–602. PDF version here.
H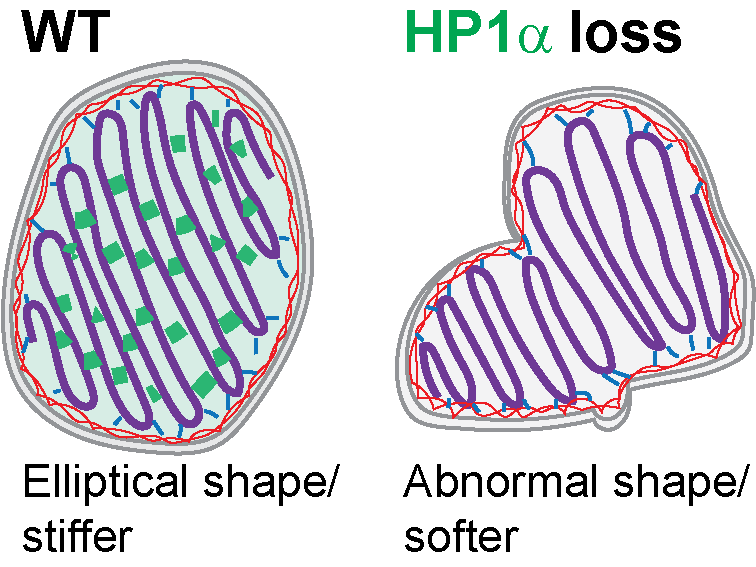 P1α is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. Strom AR, Biggs RJ, Banigan EJ, Wang X, Chiu K, Herman C, Collado J, Yue F, Ritland Politz JC, Tait LJ, Scalzo D, Telling A, Groudine M, Brangwynne CP, Marko JF, Stephens AD. (2021) eLife , 10:e63972. PDF version here.
P1α is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. Strom AR, Biggs RJ, Banigan EJ, Wang X, Chiu K, Herman C, Collado J, Yue F, Ritland Politz JC, Tait LJ, Scalzo D, Telling A, Groudine M, Brangwynne CP, Marko JF, Stephens AD. (2021) eLife , 10:e63972. PDF version here.
Chromatin and Nuclear Biophysics. Stephens AD. (2021) in press Encyclopedia of Biochemistry 3rd Edition 00272.
Liquid chromatin Hi-C characterizes compartment-dependent chromatin interaction dynamics. Belaghzal H, Borrman T, Stephens AD, Lafontaine DL, Venev SV, Weng Z, Marko JF, Dekker J. (2021) Nat Genet 53, 367. https://doi.org/10.1038/s41588-021-00784-4.
Advances in Chromatin and Chromosome Research: Perspectives from Multiple Fields. Agbleke AA, Amitai A, Buenrostro JD, Chakrabarti A, Chu L, Hansen AS, Koenig KM, Labade AS, Liu S, Nozaki T, Ovchinnikov S, Seeber A, Shaban HA, Spile JH, Stephens AD, Su JH, Wadduwage D. (2020). Mol Cell, 79, 6.
Modeling of Cell Nuclear Mechanics: Classes, Components, and Applications. Hobson CM, Stephens AD. (2020). Cells, 9, 1623. PDF version here
Chromatin rigidity provides mechanical and genome protection. Stephens AD. (2020). Mutation Research, 821, 111712. PDF preprint here
Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics. Hobson CM, Kern M, O'Brien ET, Stephens AD, Falvo MR, Superfine R. (2020). Mol Biol Cell. 31(16):1788-1801. PDF version here
High-throughput gene screen reveals modulators of nuclear shape. Tamashunas AC, Tocco VJ, Matthews J, Zhang Q, Atanasova KR, Paschall L, Pathak S, Ratnayake R, Stephens AD, Luesch H, Licht JD, Lele TP. (2020). Mol Biol Cell. 2020;31(13):1392-1402. doi:10.1091/mbc.E19-09-0520. PDF version here
Multimodal interferometric imaging of nanoscale structure and macromolecular motion uncovers UV induced cellular paroxysm. Gladstein S, Almassalha LM, Cherkezyan L, Chandler JE, Eshein A, Eid A, Zhang D, Wu W, Bauer GM, Stephens AD, Morochnik S, Subramanian H, Marko J, Ameer GA, Szleifer I, Backman V. (2019). Nat Commun. 10(1) 1652.
Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Stephens AD*, Liu PZ*, Kandula V, Chen H, Herman C, Almassahla LM, O’Halloran T, Backman V, Adam S, Goldman R, Banigan EJ, Marko JF. (2019). * co-first authors. Mol Biol Cell E19-05-0286-T. PDF version here
Chromatin’s physical properties shape the nucleus and its functions. Stephens AD, Banigan EJ, Marko JF. (2019) Curr Opin Cell Biol. 58:76-84. PDF version here
Effects of altering histone post-translational modifications on mitotic chromosome structure and mechanics. Biggs R, Liu P, Stephens AD, Marko FJ. (2019). Mol Biol Cell. 30:820-827. PDF version here
 Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Stephens AD, Liu PZ, Banigan EJ, Almassahla LM, Backman V, Adam S, Goldman R , Marko JF. (2018). Mol Biol Cell. 29: 220-233. PDF version here
Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Stephens AD, Liu PZ, Banigan EJ, Almassahla LM, Backman V, Adam S, Goldman R , Marko JF. (2018). Mol Biol Cell. 29: 220-233. PDF version here
*** We were also highlighted and got the cover!
Separate roles for chromatin and lamins in nuclear mechanics. Stephens AD, Banigan EJ, Marko JF . (2018). Nucleus. Dec 28:1-6. PDF version here
Mechanics and buckling of biopolymeric shells and cell nuclei. Banigan EJ, Stephens AD, Marko JF. (2017). Biophys J. 8: 1654-1663. PDF version here
Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Stephens AD, Banigan EJ, Adam S, Goldman R, Marko JF. (2017). Mol Biol Cell. 28: 1984-1996. PDF version here
***A list of publications and citations can be found on my Google Scholar page.
