Research interests and methods
RootFlowRT v2.8: Biological Motion Estimation for Plant Root GrowthThis material is based, partially, upon work supported by the National Science Foundation under Grant No. 031687 to TIB. Any opinions, findings, and conclusions or recommendations expressed in this material are those of Baskin and do not necessarily reflect the views of the National Science Foundation. ABSTRACT
|
||
|
|
|
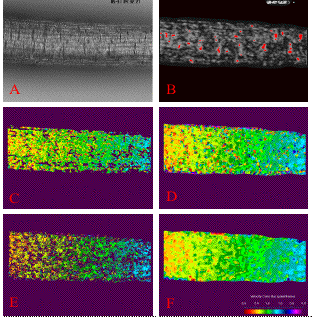
Figure 1. The mosaiced plant root is shown in the upper panel, and registered velocity profile along its medial axis in the lower panel. The velocity is relative to the base of the plant root. The corresponding 2D velocity fields used to derive the profile is shown in Figure 2. |
||
|
||
| Figure 2. The original image sequence for the third segment in the plant root mosaic shown in Figure 1 and the motion fields after each specific processing step. A. The original plant root image in a 9-frame image sequence, only frame 5 is shown; B. The motion mask in the tensor motion estimation method. C, D, E, F: the motion fields after the tensor method, the robust matching method, forward-backward consistency test, and interpolation. Only the horizontal components of the motion are displayed for each step. The vertical components of the motion are not shown here because they are very small and dominated by the noise. | ||
REFERENCING SOFTWARE RootflowRT was written by Hai Jiang and Prof. K. Palaniappan, University of Missouri-Columbia, in collaboration with Prof. Tobias I. Baskin, University of Massachusetts-Amherst. The executable code is freeware; however, in any publications that result from your using it, we ask that you send one of us a reprint, and cite the following references: van der Weele CM, Jiang H, Palaniappan KK, Ivanov VB, Palaniappan K, Baskin TI (2003) A new algorithm for computational image analysis of deformable motion at high spatial and temporal resolution applied to root growth: Roughly uniform elongation in the meristem and also, after an abrupt acceleration, in the elongation zone. Plant Physiology, 132:1138-1148. Jiang HS, Palaniappan K, Baskin TI (2003) A combined matching and tensor method to obtain high fidelity velocity fields from image sequences of the non-rigid motion of plant root growth In MH Hamza ed, IASTED International Conference on Biomedical Engineering, BioMED 2003, ACTA Press, Calgary, Canada, In press.
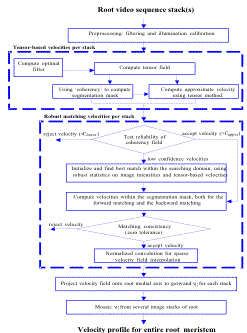
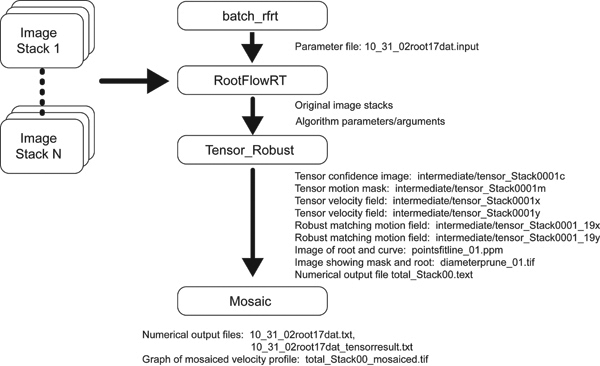
RootflowRT is a package to estimate the spatial velocity profile of plant roots. A velocity profile arises from the expansion (growth) of the elements of the root, and quantification of the velocity profile is sufficient to characterize growth fully. In principle, expansion is three-dimensional, however the software works in two dimensions only, estimating velocity in the plane perpendicular to the imaging axis (focal plane). Roots that undulate in and out of the focal plane are not suitable for analysis at present. Velocity is a vector quantity with a rate and a direction. In view of the fact that the longitudinal axis of the root provides a native axis, velocities are reported parallel and perpendicular to the local tangent to root's midline. The velocity estimation is done on an image volume obtained by capturing a set of 9 images (frames) of the growing root with the same time interval between each frame. Typical time intervals are between 2 to 30 seconds. Velocity estimation uses a structure-tensor method followed by a robust-matching method. Details are provided in the above references, our web page, or by request from us. Both methods rely on the gray level sub-structure (i.e. texture) within the root image. Because elongation is typically of chief interest, the numerical velocity profiles are created by averaging velocity the locus of values perpendicular to each point of the root's midline. The midline can be obtained either algorithmically by the software or can be manually specified by the user. The latter method is very useful for complex roots with many root hairs or other incidental features of the image that make automatic root segmentation more error prone and prevent the program from being 100% successful. To pick midline points, obtain the coordinates of points near the left and right edges of the root, and if the root is bent, then include one or two additional points along the root. Input images must be in either TIFF or PPM format and are typically 640 pixels by 480 pixels and 8 bits per pixel. Support for larger image sizes such as 1280 x 1024, as well as higher gray level quantization resolution of 12 bits per pixel is being added to the software. Each set of 9 images (or frames) is called a stack and the software expects the default filenames of, tiff/stack####frame####, where tiff is the subdirectory name and the # symbol represents one numeric digit and there is a space after the word Frame. Hence up to 9999 stacks each with 9999 frames can be accommodated using this naming scheme; however, the software currently handles up to 99 stacks. In addition to the input image sequences, an input text file is required to specify various parameters for the algorithm. As output, for each segment, the software produces a set of images to show intermediate results and tabulates data for each segment, and for the complete profile produces two sets of tabulated results and a graph. Since the root is long and thin, and larger than the field of view of the microscope for typical resolutions, a set of stacks (or segments) must be obtained, which when mosaiced together span the growth zone. Overlaps between stacks should be about 20 % of the field of view. RootflowRT calculates the velocity profile for each segment and mosaics them together for a complete profile. Presently, two kinds of mosaic can be handled. In the simplest way, the user takes a 10th image after the 9 in the stack of a highly textured background. The background image file name must be specified by convention as stack####back0001. The focus can be changed to capture the background image, but not the x, y stage position. The software will then mosaic the profiles from each stack by finding the overlap between background images which should have some distinct textural components for the algorithm to produce reliable results. Alternatively, if the user can translate the stage by a specific distance, the software can mosaic the profiles from this information and background images do not need to be obtained. The complete profile is also transformed so that the quiescent center (or root tip) is the origin (i.e., spatial coordinate of zero, velocity of zero). This is the natural frame of reference for the root. The velocity is transformed by averaging values in the vicinity of the tip and subtracting the measured values from these. This can create negative values in places in the profile. For transforming the distance coordinate, the movement of the tip between stacks is accounted for using a steady state growth model with constant tip velocity. RootflowRT was developed on SGI workstations under IRIX 6.5 and has been ported to Windows(98, 2000, NT), Mac OSX, and Linux(RedHat). The software is written in ANSI C. This software can be installed into ~username/bin or system wide, as long as the user's command path includes the command directory with the executable programs. For MS Windows, the user can download cygwin from http://www.cygwin.com to emulate the Unix environment, following the Windows Version Readme to install the Windows version. Before running the software, the user needs to create a working directory to manage the imagery acquired from the video microscope and the plethora of outputs produced by RootflowRT. The user can choose any name, accepted by the operating system, for the working directory. The working directory contains four sub-directories, pgm, intermediate, curve and tiff. The original root image sequences are expected by default to be in the sub-directory tiff. RootflowRT can also read input image sequences in the PPM format if the images are placed in the pgm sub-directory. The default name of the root images are tiff/stack####frame#### such as tiff/stack0001frame0001. Note that the input image stack consists of a set of 9 separate files, with frame 5 being the center frame. In addition to the image sequences, the user also needs to provide parameter values for the run. Although this data can be entered manually as the program runs or as command line input to the batch programs, it is generally more convenient to organize the user responses into a single input specification file that can be read by the program as needed using the Unix file redirection feature. The format of the input file is given next. The software will ask a series of questions, and the answers can be saved into an input file. The lines of the input file contain either general parameters (such as the camera calibration to convert pixels to microns), specific parameters such as the number of segments for the root, and switches for certain choices of how the software runs. The syntax of this file is rigid since it replaces manual entry by the user and a sample file is given below. Following that is a set of definitions or explanations for each line. Sample Input Data File: ----------------------------------------------------------------------------------------------------------------- If the user does not specify the geometry input for one stack, by omitting the lines for stack 3 in the input file then the program will generate the medial axis and diameters for stack 3 from the motion mask itself, but will use the user input midline points for the other stacks. If no geometrical information is given then all stacks will be automatically handled by the program. The data processing flow chart below shows the order in which the three programs comprising the suite of RootFlowRT software, need to be run along with the output files produced by each program. The simple command script, batch_rfrt, controls the order of execution of, rootflowrt, tensor_robust and plot_mosaic. The user only needs to run the Unix shell command script, batch_rfrt: |
||
 |
||
batch_rfrt [working directory] < 10_31_02root17dat.input where input.txt is the input parameter file and the less than symbol, < is the Unix file redirection command. If the user is in the working directory then the Unix shorthand for the current working directory can be used: batch_rfrt . < 10_31_02root17dat.input The command script batch_rfrt simply specifies which root sequence in the tiff sub-directory of the working directory to use. The script consists of just two commands: cd [work directory] assuming the default root image names are used, Stack####Frame #### where # is a numeric digit. This script is easily modified to use other file names or to adapt to the local directory environment on different computing platforms. The user can run the software successfully with the above information, however, more advanced usage can be found in the advanced information section. ADVANCED INFORMATION (Command Line Arguments) RootflowRT is a program to parse the input parameter file, launch the motion estimation program tensor_robust for each image stack, then finally call plot_mosaic to generate the complete mosaiced velocity profiles. The program rootflowrt reads the input file and translates parameters into command line arguments for tensor_robust and plot_mosaic . A novice user can launch the command rootflowrt by using batch_rfrt , for the details please read the usage of batch_rfrt . For the expert user, the command line syntax for tensor_robust is as follows: A sample usage of tensor_robust is: The command line arguments for plot_mosaic is: A sample usage of plot_mosaic is: The program rootflowrt will print out the command line with the specification in the input file when it launches tensor_robust and plot_mosaic . So the user can observe what are passed to the two commands. DOWNLOAD THE SOFTWARE Please submit your contact information before downloading RootflowRT We collect your contact information only for better research cooperation and getting feedback from you for the software. Your contact information will not be released to third party people or organizations. |
||